Inverse analysis of QSPR using mol-infer
- Prediction of molecular structure from physical property values
- Inverse analysis using chemical graphs
- Application to molecular design targeting partition coefficients as desired properties
Prediction of molecular structure from physical property values
Data for a partition coefficient (logP = 10.0) set as the target property for inverse analysis using mol-infer are shown. First, data for 1,297 molecular structures and property values are used to train an artificial neural network (ANN) on the structure–property relationship. Inverse analysis is then performed using the trained model.
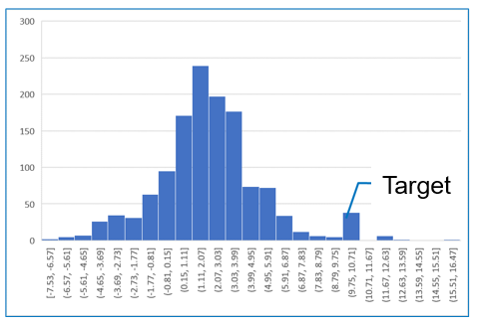
Inverse analysis using chemical graphs
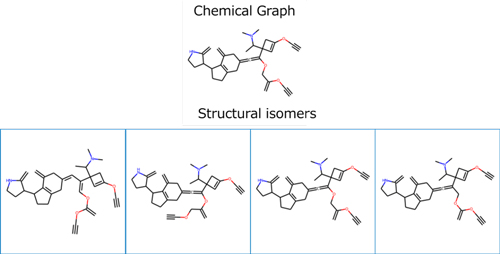
The seed structures of molecular structures used for inverse analysis and tree structures corresponding to functional groups are shown. By combining these structural elements using mixed-integer linear programming (MILP), molecular structures that satisfy target properties are searched.
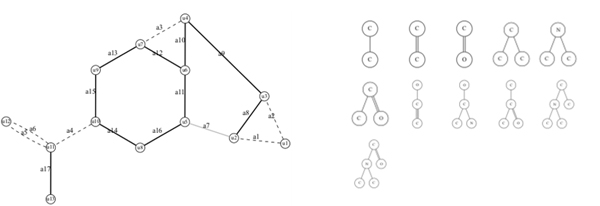
Application to molecular design targeting partition coefficients as a property
An example of molecular structures obtained by inverse analysis is shown. Structural isomers are also included, and forward property estimation was performed again for the obtained structures. The partition coefficient was 9.8, close to the target value, confirming the effectiveness of the inverse analysis.